The Journal of Advances in Parasitology
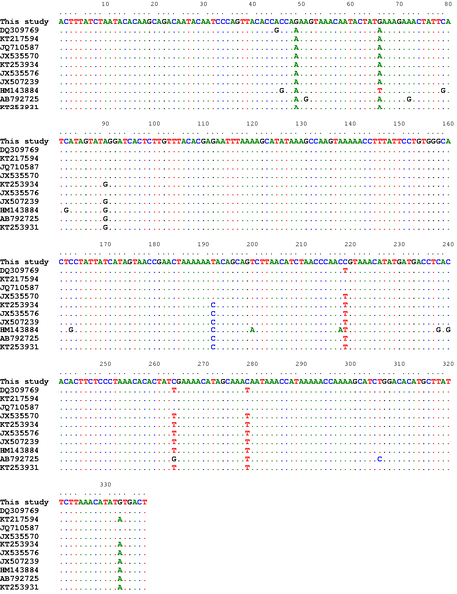
Alignment of the CO1 partial sequences (338bp) of our isolates with a number of T. multiceps isolates from variable host species within different geographical regions including sheep (DQ309769, KT217594, JQ710587, JX535570), goats (KT253934, JX535576, JX507239), cattle (HM143884) and dogs (AB792725, KT253931)
Alignment of the ND1 partial sequences (419bp) of our isolates with a number of T. multiceps isolates from variable host species within different geographical regions including sheep (DQ309770, DQ077820, KF233942, AJ239104, KT253935), goats (KT253936, KC794811), cattle (HM143887) and dogs (KT253932)
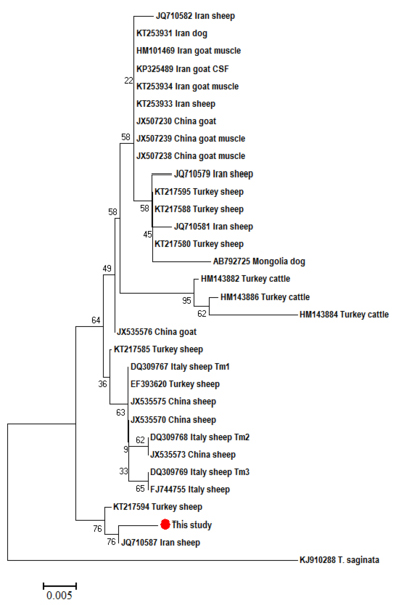
Neighbor-Joining phylogenetic tree of T. multiceps CO1 partial sequences from different hosts and geographical regions. T. saginata was used as an out group. Sequences were aligned by Bioedit and the tree was built using the software MEGA (version 6) with the Kimura 2 parameter mode. Scale bar indicates the proportion of sites changing along each branch
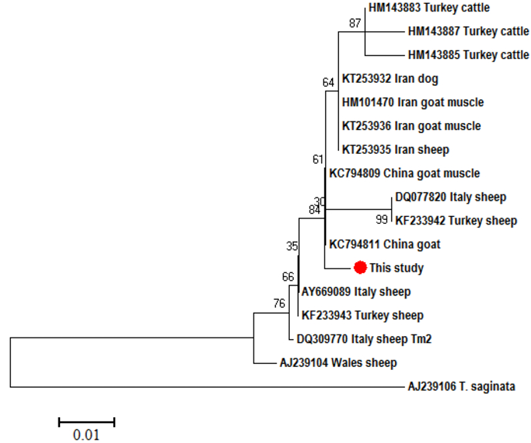
Neighbor-Joining phylogenetic tree of T. multiceps ND1 partial sequences from different hosts and geographical regions. T. saginata was used as an out group. Sequences were aligned by Bioedit and the tree was built using the software MEGA (version 6) with the Kimura 2 parameter mode. Scale bar indicates the proportion of sites changing along each branch